
Our lab’s vision is to better understand brain functions and its related disorders by designing specialized interpretable tools from machine learning and statistics. More specifically, our research programs revolve around the development of transparent supervised and unsupervised machine learning tools to integrate multi-modal data collected from brain (and body) in both microscopic and macroscopic resolutions. Some of the specific projects in the lab include:
Functional modeling in the brain
We develop models and algorithms based on advanced machine learning principles to understand the functions of neurons in the brain and their relationship with spatially resolved gene expression profiles. To achieve this, we study large-scale datasets with a variety of modalities such as electrophysiology, calcium imaging, electron microscopy, and spatial transcriptomic and gene expression profiles.
Specific projects:
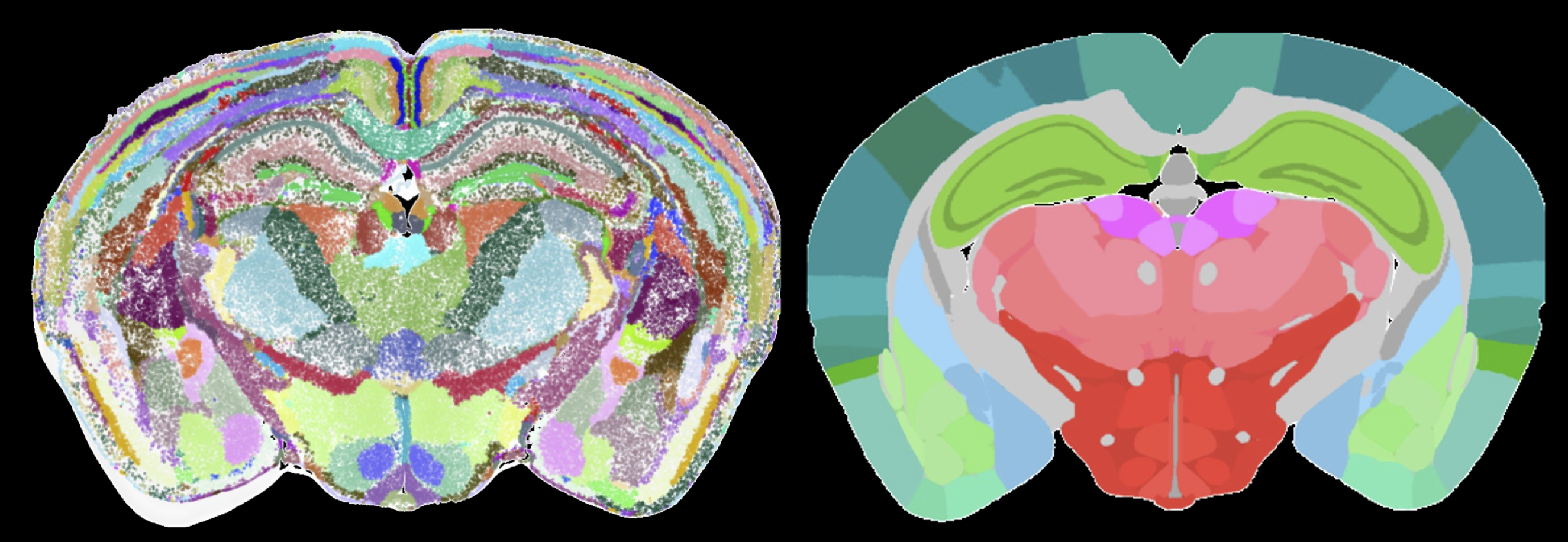
Defining the function of any organ including the brain depends on defining and describing its building blocks: tissues, tissue domains, and cell types. Genome-wide characterization of gene expression and machine learning approaches have transformed the understanding of cell types that build the nervous system. However, the precise arrangement of cells types and their differences across the brain areas will require interpretation of spatial gene expression data. In collaboration with Hongkui Zeng and Bosiljka Tasic at the Allen Institute and Bin Yu at UC Berkeley, this project aims to integrate spatial gene expression and neural connectivity data to reveal building blocks of spatial gene expression profiles in the mouse and human brain. These building blocks will partition the brain into completely data-driven functional 3D brain areas and establish local gene networks. The project involves designing and validating unsupervised and interpretable machine learning frameworks and statistical tools.
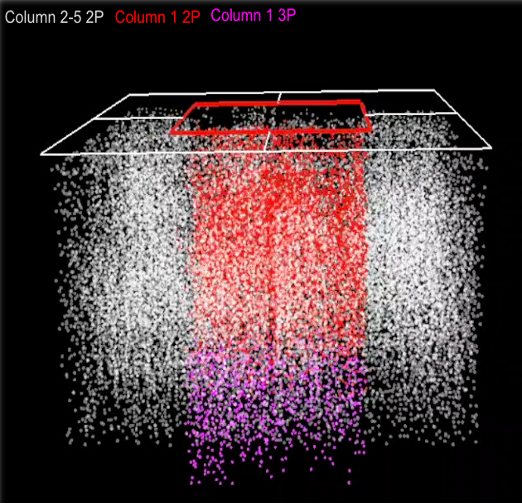
In another project supported by an R01 grant from NIH/NIMH, we aim to characterize the relationship between neural function and connectivity in visual sensory processing. In order to explore this relationship and in collaboration with the Allen Institute, we are analyzing recorded visual responses from pan-excitatory neurons within the 1 mm cube region of the primary visual cortex, spanning all visual layers from pia to white matter. This includes 750 2-photon and 35 3-photon calcium imaging planes spaced by ~16 um. Our goal is to examine the single-cell and population activity in the primary visual cortex integrated with Electron Microscopic reconstruction of neural morphology and connectivity from the same tissue to understand the principles of functional connectivity at the single-cell level.
Computational clinical neuroscience
As a part of the UCSF Neuroscape Center and in order to bridge our understanding of the brain to patient data, we build tools to study and visualize large-scale neurological clinical datasets. Our goals are to characterize the relationship between multi-modal patient data and disease processes to enable efficient biomarker identification. This involves analyzing and visualizing longitudinal medical images, genomic data, biosensor data, and other relevant datasets collected from patients.
Specific projects:

Multiple Sclerosis (MS) is the leading cause of nontraumatic neurological disability in young adults, and it is estimated that 3 in 1000 US adults, or nearly 1 million individuals, are affected. The development of effective therapeutics for MS represents one of the great success stories of modern molecular medicine, with near-complete suppression of clinical attacks and focal brain inflammation now possible for most patients. However, the more disabling neurodegenerative component of the disease – progressive MS – remains poorly treated, and the development of potent therapeutics for progressive MS has been limited by an inability to efficiently extract clinically meaningful MRI data corresponding to changes in white matter lesions, global and regional atrophy, and neurodegeneration. Various data modalities are used to assess the progression of MS including brain MRIs, behavioral measures, and genomic data. The multi-modal nature of the large-scale data collected from these patients demand for automated computational tools processing methods. In collaboration with the UCSF Multiple Sclerosis (MS) Center, this project aims to systematically integrate and analyze longitudinal MRI and genomics data from hundreds of patients with MS to better understand the disease process and its biomarkers. More specifically, this project leverages one-of-a-kind UCSF EPIC and ORIGINS MS datasets to build state-of-the-art interpretable machine learning pipelines to characterize dynamic changes over time and their relationship to MS progression. Additionally, the project involves the development of state-of-the-art and transparent machine learning tools to build computational pipelines for 3T and 7T MRI.

A deeper understanding of an individual’s state (e.g, stress, mood, attention, arousal, awareness) requires recording continuous data across multiple modalities and integrating these signals to generate meaningful and predictive composite measures. In collaboration with the UCSF Neuroscape Center, this project aims to systematically integrate hundreds of different biosensor data collected from the brain and body to predict the emotional state in humans. The data consists of high-density EEG, EMG, ECG, EOG, Continuous blood pressure, Trans Radial Electrical Impedance Velocimetry, respiration, EDA/GSR, and accelerometers readings. This project involves the development of interpretable machine learning tools to integrate and analyze large-scale time-series data.

Advanced Parkinson’s disease (PD) is characterized by motor and non-motor symptoms which are highly disabling and significantly impair quality of life. Optimizing dopaminergic medication, Deep Brain Stimulation (DBS), and lifestyle interventions to minimize symptoms is central to the management of PD. Achieving this goal requires accurate measures of symptom severity and fluctuations. At-home objective symptom quantification introduces a potential solution, but due to technical limitations, wearable tracking devices have not yet been readily adopted into clinical practice for PD. In collaboration with the UCSF Movement Disorders and Neuromodulation Center and Google Research, this project aims to develop the next generation of at-home video-assisted technology to track and diagnose PD. This solution is enabled with advanced machine learning tools and has the potential to significantly enhance diagnostic accuracy and optimize personalized medical therapy in PD.
Interpretable Machine Learning
Guided by scientific questions from neuroscience and biomedicine, we are interested in the general problem of interpreting machine learning models. In the past decade, research in machine learning has been principally focused on the development of algorithms and models with high predictive capabilities. However, interpreting these models remains a challenge, primarily because of the large number of parameters involved. We investigate methods based on statistical principles to build more interpretable machine learning models.